import time
import numpy as np
import scipy
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import paxplot
import pickle
from functools import partialIn [1]:
In [2]:
with open("data/sim_long_preprocessed_slice.pkl", "rb") as file:
import_dict = pickle.load(file)
import_dict.keys()dict_keys(['parameters', 'patterns'])In [3]:
test_pattern = import_dict["patterns"][0]#[::2, ::2]
plt.imshow(test_pattern)
test_pattern.shape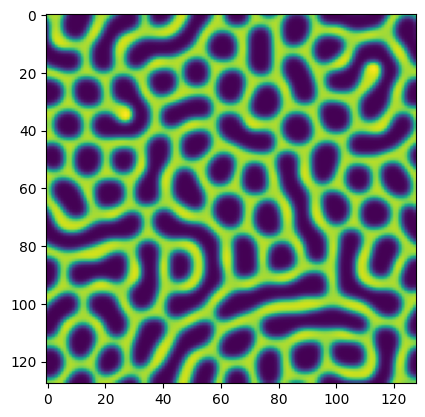
In [4]:
def get_adj_matrix_slow(pattern, eps=0.003):
assert pattern.shape[0] == pattern.shape[1]
graph_res = pattern.shape[0]
m = graph_res**2
adj_matrix = np.zeros((m, m), dtype=np.float64)
u_mean = np.mean(pattern)
for i in range(m):
neighbor_js = np.array([i - 1, i + 1, i - graph_res, i + graph_res])
neighbor_js = neighbor_js % m
for j in neighbor_js:
u_vi = pattern[i % graph_res, i // graph_res]
u_vj = pattern[j % graph_res, j // graph_res]
vi_high = u_vi > u_mean
vj_high = u_vj > u_mean
if not vi_high ^ vj_high: # both on same side of u_mean
adj_matrix[i, j] = 1
else:
adj_matrix[i, j] = eps
return adj_matrix
def compute_resistance_slow(pattern, eps):
assert pattern.shape[0] == pattern.shape[1]
graph_res = pattern.shape[0]
m = graph_res**2
adj_matrix = get_adj_matrix_slow(pattern, eps)
graph_laplacian = np.diag(adj_matrix @ np.ones((m,))) - adj_matrix
K = scipy.linalg.inv(np.ones((m, m)) + graph_laplacian)
Kvivi = np.ones((m, m)) * np.diagonal(K)[:, None]
Kvjvj = Kvivi.T
resistance = Kvivi + Kvjvj - 2 * K
assert resistance.shape == (m, m)
return resistance
eps = 0.003t1 = time.time() res_dist_reference = compute_resistance_slow(test_pattern, eps); time_reference = time.time() - t1 f”took {time_reference:.4f}s | mean={np.mean(res_dist_reference):.4f}”
In [5]:
fig, ax = plt.subplots(figsize=(8,8), dpi=100)
adj_matrix = get_adj_matrix_slow(test_pattern, 0.5)
m = adj_matrix.shape[0]
Z = graph_laplacian = np.diag(adj_matrix @ np.ones((m,))) - adj_matrix
extent = (0,m,0,m)
divnorm=mpl.colors.TwoSlopeNorm(vcenter=0., vmin=Z.min(), vmax=Z.max())
img = ax.imshow(Z, origin="upper", extent=extent, cmap="RdBu", norm=divnorm)
ticks = [0, 1*m//4, m//2, 3*m//4, m]
ax.set_xticks(ticks)
ax.set_xticklabels(ticks)
ax.set_yticks(ticks)
ax.set_yticklabels(ticks[::-1])
crop_size = 200
# inset Axes....
x1, x2, y1, y2 = 0,crop_size,m-crop_size,m # subregion of the original image
axins = ax.inset_axes(
[0.3, 0.2, 0.9, 0.9],
xlim=(x1, x2), ylim=(y1, y2), xticklabels=[], yticklabels=[])
axins.imshow(Z, origin="upper", extent=extent, cmap="RdBu", norm=divnorm)
ax.indicate_inset_zoom(axins, edgecolor="black", lw=2)
plt.show()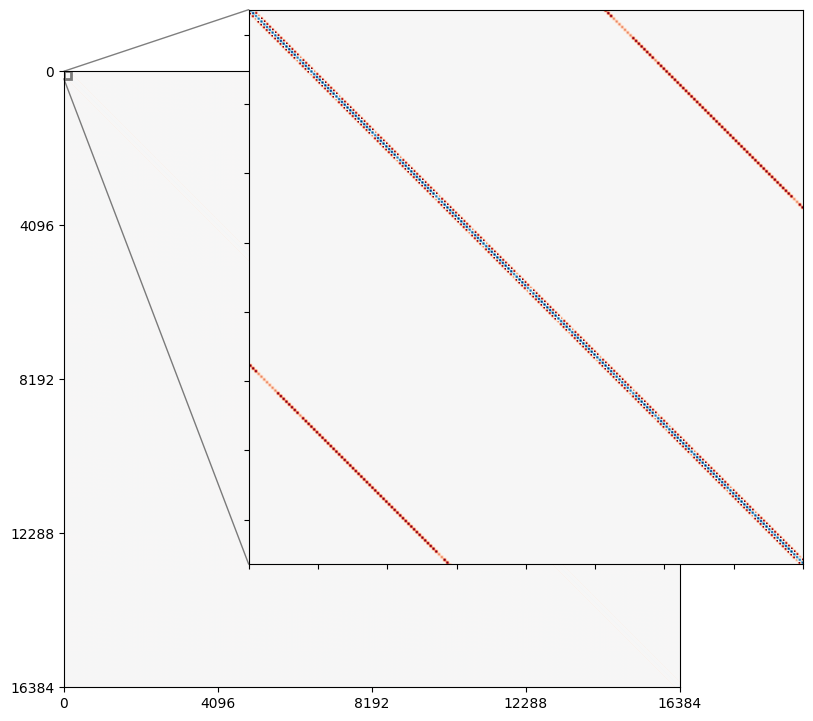
In [6]:
test_pattern = import_dict["patterns"][0][:30:2, :30:2]In [7]:
graph_res = test_pattern.shape[0]
m = graph_res**2
adj_matrix = get_adj_matrix_slow(test_pattern, eps)
graph_laplacian = np.diag(adj_matrix @ np.ones((m,))) - adj_matrix
K = scipy.linalg.inv(np.ones((m, m)) + graph_laplacian)
Kvivi = np.ones((m, m)) * np.diagonal(K)[:, None]
Kvjvj = Kvivi.T
plt.imshow(Kvjvj)
plt.colorbar()
In [8]:
t1 = time.time()
res_dist = compute_resistance_slow(test_pattern, eps);
time_reference = time.time() - t1
f"took {time_reference:.4f}s" # | mean={np.mean(res_dist_reference):.4f}"'took 0.1246s'In [9]:
plt.imshow(res_dist)
plt.colorbar();
In [10]:
def prepare_graph_laplacian_matrix(pattern, eps):
graph_res = pattern.shape[0]
m = graph_res**2
adj_matrix = get_adj_matrix_slow(pattern, eps)
graph_laplacian = np.diag(adj_matrix @ np.ones((m,))) - adj_matrix
return graph_laplacian
laplacian = prepare_graph_laplacian_matrix(test_pattern, eps)
# K = scipy.linalg.inv(np.ones((m, m)) + graph_laplacian)In [11]:
%%timeit
scipy.linalg.pinv(laplacian)142 ms ± 6.22 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)In [12]:
generalized_inverse = scipy.linalg.pinv(laplacian)In [13]:
def check_g_inverse(A, G):
return np.isclose(A @ G @ A, A).mean() == 1.0
check_g_inverse(laplacian, generalized_inverse)TrueIn [14]:
def benchmark_g_inv_fun(A, fun, num_loops=3):
t1 = time.time()
for i in range(num_loops):
G = fun(A)
t_total = time.time() - t1
assert check_g_inverse(A, G), "The provided function did not compute a correct g-inverse."
return t_total / num_loopsIn [15]:
laplacian.shape(225, 225)In [16]:
benchmark_g_inv_fun(laplacian, np.linalg.pinv)0.025146484375In [17]:
benchmark_g_inv_fun(laplacian, scipy.linalg.pinv)0.13741509119669595In [18]:
def imqrginv(a: np.ndarray) -> np.ndarray:
q, r = np.linalg.qr(a, mode="reduced")
return r.transpose(1, 0).conj() @ np.linalg.inv(r @ r.transpose(1, 0).conj()) @ q.transpose(1, 0).conj()
benchmark_g_inv_fun(laplacian, np.linalg.pinv)0.038936456044514976In [19]:
def add_ones_before_fun(A, fun):
return fun(np.ones_like(A) + A)
maybe_g_inv = add_ones_before_fun(laplacian, np.linalg.inv)
assert check_g_inverse(laplacian, maybe_g_inv)In [20]:
benchmark_g_inv_fun(laplacian, partial(add_ones_before_fun, fun=np.linalg.inv))0.0011440118153889973In [21]:
benchmark_g_inv_fun(laplacian, partial(add_ones_before_fun, fun=np.linalg.pinv))0.009821097056070963In [22]:
g_inv_funs = {
"+1-np-inv": partial(add_ones_before_fun, fun=np.linalg.inv),
"+1-scipy-inv": partial(add_ones_before_fun, fun=scipy.linalg.inv),
"np-pinv": np.linalg.pinv,
"scipy-pinv": scipy.linalg.pinv,
"np-pinvh": partial(np.linalg.pinv, hermitian=True),
"scipy-pinvh": scipy.linalg.pinvh,
}
results_small_m = {}
for pattern_size in [8, 16, 32, 64]:
test_pattern = import_dict["patterns"][0][:pattern_size, :pattern_size]
laplacian = prepare_graph_laplacian_matrix(test_pattern, eps)
m = laplacian.shape[0]
print(f"graph has {m} vertices.")
results_small_m[m] = {}
for key, fun in g_inv_funs.items():
if "+1" in key: # repeat the faster algorithms a bit more often to reduce sampling error
if pattern_size < 30:
num_loops = 50
else:
num_loops = 20
else:
if pattern_size < 30:
num_loops = 20
else:
num_loops = 8
elapsed = benchmark_g_inv_fun(laplacian, fun, num_loops=num_loops)
print(elapsed, key)
results_small_m[m][key] = elapsedgraph has 64 vertices.
0.00011413097381591797 +1-np-inv
0.00014008522033691405 +1-scipy-inv
0.0012161016464233398 np-pinv
0.008746886253356933 scipy-pinv
0.002484130859375 np-pinvh
0.0036941170692443848 scipy-pinvh
graph has 256 vertices.
0.00465179443359375 +1-np-inv
0.00352142333984375 +1-scipy-inv
0.025093352794647215 np-pinv
0.16102132797241211 scipy-pinv
0.019651973247528078 np-pinvh
0.18433376550674438 scipy-pinvh
graph has 1024 vertices.
0.05806041955947876 +1-np-inv
0.06515189409255981 +1-scipy-inv
0.575680673122406 np-pinv
0.7435911595821381 scipy-pinv
0.20940321683883667 np-pinvh
0.9509794414043427 scipy-pinvh
graph has 4096 vertices.
2.1050000548362733 +1-np-inv
3.566044104099274 +1-scipy-inv
31.111353754997253 np-pinv
34.37004932761192 scipy-pinv
13.204486846923828 np-pinvh
69.60303673148155 scipy-pinvhIn [23]:
import warnings
warnings.filterwarnings("ignore", message="^.*is not officially supported by Paxplot, but it may still work.*$")In [24]:
res_df = pd.DataFrame(results_small_m)
cols = res_df.columns
col_names = ["m="+str(col) for col in cols]
# Create figure
paxfig = paxplot.pax_parallel(n_axes=len(cols))
paxfig.plot(res_df.to_numpy()[:2,], line_kwargs={"linestyle": "-"})
paxfig.plot(res_df.to_numpy()[2:4], line_kwargs={"linestyle": "--"})
paxfig.plot(res_df.to_numpy()[-2:], line_kwargs={"linestyle": ":"})
paxfig.set_labels(col_names)
plt.suptitle("Elapsed time [s]", y=1.01)
# Changing figure size
paxfig.set_size_inches(5, 3)
paxfig.set_dpi(200)
paxfig.subplots_adjust(left=0.1, bottom=0.2, right=0.9, top=0.9) # Padding
# Change tick size
tick_size = 10
for ax in paxfig.axes:
ax.tick_params(axis="y", labelsize=tick_size)
# Change label size
label_size = 15
for ax in paxfig.axes:
ax.tick_params(axis="x", labelsize=label_size)
for i, m in enumerate(cols):
max_tick = res_df[m].max()
paxfig.set_lim(i, 0, max_tick * 1.01)
paxfig.set_even_ticks(
ax_idx=i,
n_ticks=4,
minimum=0,
maximum=max_tick,
)
lines = paxfig.axes[0].lines
labels = [i for i in res_df.index]
plt.legend(lines, labels, bbox_to_anchor=(1.5, 1),
loc='upper left', borderaxespad=3.5);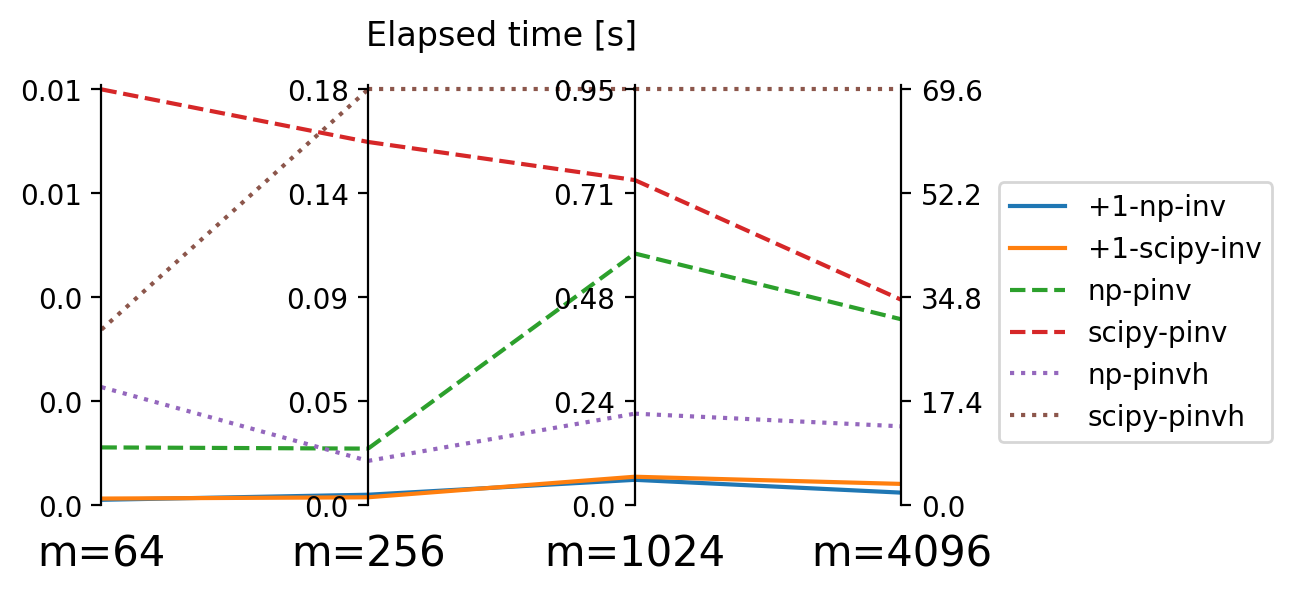
numpy and scipy accross matrix size \(m\).
In [25]:
g_inv_funs = {
"+1-np-inv": partial(add_ones_before_fun, fun=np.linalg.inv),
"+1-scipy-inv": partial(add_ones_before_fun, fun=scipy.linalg.inv),
}
results_large_m = {}
for pattern_size in [16, 32, 64, 128]:
test_pattern = import_dict["patterns"][0][:pattern_size, :pattern_size]
laplacian = prepare_graph_laplacian_matrix(test_pattern, eps)
m = laplacian.shape[0]
print(f"graph has {m} vertices.")
results_large_m[m] = {}
for key, fun in g_inv_funs.items():
elapsed = benchmark_g_inv_fun(laplacian, fun, num_loops=3)
print(elapsed, key)
results_large_m[m][key] = elapsedgraph has 256 vertices.
0.0024029413859049478 +1-np-inv
0.041563828786214195 +1-scipy-inv
graph has 1024 vertices.
0.546661376953125 +1-np-inv
0.06827251116434734 +1-scipy-inv
graph has 4096 vertices.
2.0935885111490884 +1-np-inv
1.9664779504140217 +1-scipy-inv
graph has 16384 vertices.
82.14190308252971 +1-np-inv
99.59056385358174 +1-scipy-invIn [26]:
res_df = pd.DataFrame(results_large_m)
cols = res_df.columns
col_names = ["m="+str(col) for col in cols]
# Create figure
paxfig = paxplot.pax_parallel(n_axes=len(cols))
paxfig.plot(res_df.to_numpy(), line_kwargs={"linestyle": "-"})
paxfig.set_labels(col_names)
plt.suptitle("Elapsed time [s]", y=1.01)
# Changing figure size
paxfig.set_size_inches(5, 3)
paxfig.set_dpi(200)
paxfig.subplots_adjust(left=0.1, bottom=0.2, right=0.9, top=0.9) # Padding
# Change tick size
tick_size = 10
for ax in paxfig.axes:
ax.tick_params(axis="y", labelsize=tick_size)
# Change label size
label_size = 12
for ax in paxfig.axes:
ax.tick_params(axis="x", labelsize=label_size)
for i, m in enumerate(cols):
max_tick = res_df[m].max()
paxfig.set_lim(i, 0, max_tick * 1.01)
paxfig.set_even_ticks(
ax_idx=i,
n_ticks=4,
minimum=0,
maximum=max_tick,
)
lines = paxfig.axes[0].lines
labels = [i for i in res_df.index]
plt.legend(lines, labels, bbox_to_anchor=(1.5, 1),
loc='upper left', borderaxespad=3.5);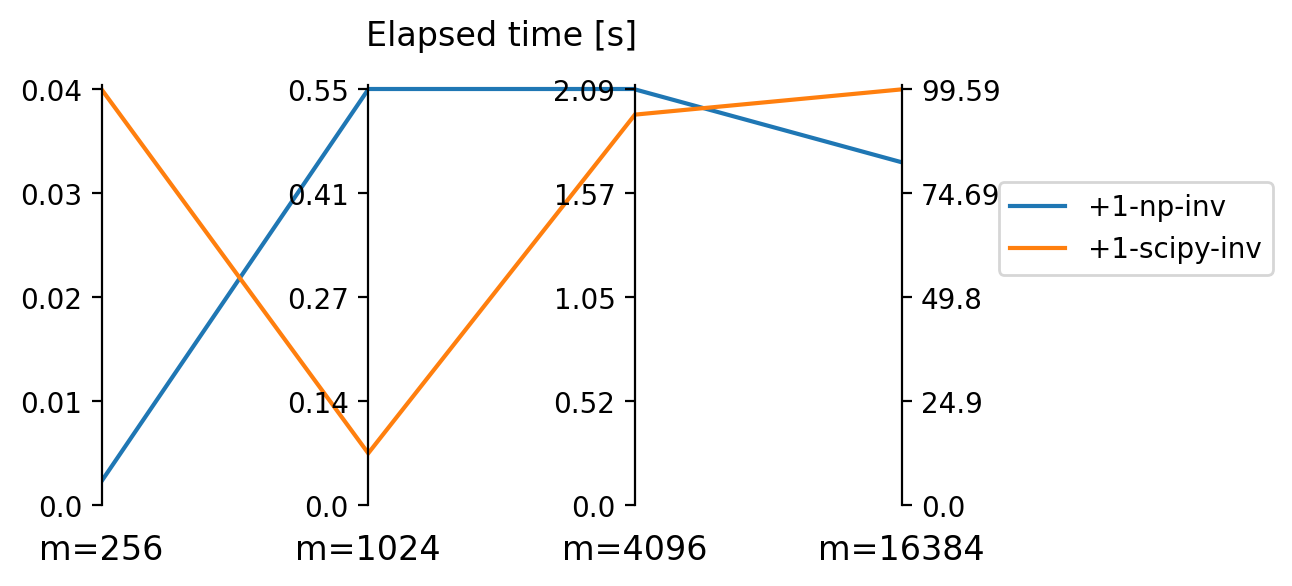
numpy and scipy for very high matrix size \(m\).